The Plant Viruses and Viral Genetic Engineering Technology Specializing Research Team from our college has reported on the cross-kingdom infection of plant viruses (CMV), viroids (HSVd), and fungal viruses (CHV1) in plants and fungi in the Proceedings of the National Academy of Sciences (PNAS) in 2017, 2019, and 2020. Recently, the team published a research paper entitled "Identification of a negative-strand RNA virus with natural plant and fungal hosts" in PNAS. This study reports a novel negative-strand RNA virus, Valsa mali negative-strand RNA virus 1 (VmNSRV1), that exhibits cross-kingdom infection in plants and fungi. Ph.D student Dai Ruoyin and Doctor Yang Shi'an who has graduated from our college are the co-first authors of this paper, while Prof. Sun Liying from our college and Prof. Ida Bagus Andika from the College of Plant Protection of Qingdao Agricultural University are the co-corresponding authors.
During long-term evolution, viruses and their hosts exhibit a certain degree of selectivity. As viruses undergo genetic mutations or changes in their transmission modes, their host range also expands. Viruses can not only cross species barriers, but a few can even cross kingdoms to infect new hosts. Studies have found that cross-species/cross-kingdom virus infections and transmissions have led to outbreaks of important viral diseases. Therefore, this phenomenon is a hot topic and frontier in virology research.
The novel fungal virus VmNSRV1 discovered in this study was isolated from the pathogenic fungus Valsa mali, which causes apple tree rot. VmNSRV1 can significantly reduce the pathogenicity of Valsa mali, making it a novel and highly effective biocontrol fungal virus. The VmNSRV1 genome contains three negative-strand RNA segments (RNA1, RNA2, RNA3), encoding the viral RNA-dependent RNA polymerase (RdRp), a movement-like protein (MP-L), and a nucleoprotein (N), respectively. Phylogenetic analysis based on the conserved structural domain of the viral RdRp revealed that VmNSRV1 is closely related to the Laulavirus genus within the Phenuiviridae family and is temporarily classified as a new member of this genus. Interestingly, VmNSRV1 RNA2 encodes an MP-L protein that can be localized to the plasmodesmata of plant cells and exhibits the function of plant virus movement proteins. The study found that VmNSRV1 can not only systematically infect plantsbut also transmit bidirectionally between plants and plant-related pathogenic fungi. Additionally, among 139 apple tree leaves tested in orchard experiments in Shaanxi Province, 24 were positive for VmNSRV1. This suggests that under natural conditions, VmNSRV1 exhibits cross-kingdom infection between plants and pathogenic fungi. Given its excellent biocontrol potential and extracellular transmission ability, the study invented a "biocontrol fungal virus" spraying technique, where fungal extracellularly secreted virus particles are directly sprayed onto infected plants, effectively inhibiting the expansion of apple tree rot lesions. Furthermore, the nucleocapsid N protein encoded by VmNSRV1 RNA3 is associated with lipid droplet metabolism in fungal and plant cells, which may be the key to its cross-kingdom infection of fungi and plants.
This study not only provides a new strategy for the biological control of plant fungal diseases but also holds significant implications for understanding the adaptability and evolution of plant and fungal viruses.
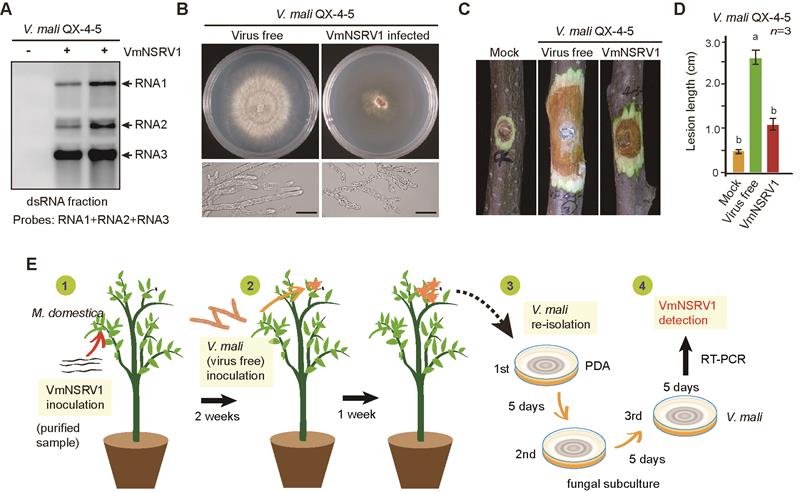
Analysis of the biological characteristics and cross-kingdom transmission of the virus VmNSRV1
Professors Kang Zhensheng and Wu Yunfeng from our college also participated in this research. The study was funded by grants from the National Natural Science Foundation of China (31970163) and the Natural Science Foundation of Xinjiang Uygur Autonomous Region (2021D01D12).
Original link: www.pnas.org/doi/10.1073/pnas.2319582121