Recently, the research group led by Prof. Liu Huiquan from Functional Genomics of Plant Pathogenic Fungi Research Team published an online paper titled Unveiling the A-to-I mRNA editing machinery and its regulation and evolution in fungi in the journal Nature Communications, revealing the enzyme complex for A-to-I mRNA editing in fungi and clarifying its origin, evolution, and regulatory mechanisms. This study provides important new insights for fungal disease prevention and control as well as the development of gene editing tools.

A-to-I mRNA editing is a crucial mechanism for modifying genetic information, converting adenosine (A) to inosine (I) in mRNA molecules. Since inosine (I) is chemically similar to guanosine (G), it is recognized and used as guanosine (G) during protein translation, potentially leading to changes in the encoded protein sequence. In the past, A-to-I mRNA editing has mainly been studied and reported in animals, but in 2016, Prof. Liu's research group first discovered this phenomenon in fungi and demonstrated its crucial role in sexual reproduction and adaptive evolution in fungi such as Fusarium graminearum. In animals, A-to-I mRNA editing is catalyzed by ADAR enzymes, but fungi do not possess homologs of ADAR genes, making the mechanism of A-to-I mRNA editing in fungi a scientific mystery. After nearly eight years of in-depth research, Professor Liu's team has finally uncovered the mechanism of A-to-I mRNA editing in fungi. The main findings are:
A-to-I mRNA editing in fungi is catalyzed by the Tad2-Tad3-Ame1 enzyme complex. The Tad2-Tad3 heterodimer is a conserved tRNA-specific adenosine deaminase in eukaryotes, primarily responsible for A-to-I editing at position 34 of the tRNA anticodon loop. A specific protein Ame1 evolved in fungi breaks this substrate recognition restriction, enabling the Tad2-Tad3-Ame1 ternary complex to recognize and edit mRNA.
The reason for the specific occurrence of A-to-I mRNA editing in the sexual reproductive stage of fungi. The mRNA editing activator Ame1 is only induced to express during the sexual reproductive stage, which is the direct reason for the specific occurrence of A-to-I mRNA editing at this stage. Additionally, transcriptional and post-translational regulations also play crucial roles in regulating the activity of the editing enzyme complex during the sexual reproductive stage. Although A-to-I mRNA editing is beneficial to fungal sexual reproduction, artificial expression of Ame1 to activate A-to-I mRNA editing during the asexual stage is detrimental to the growth and infection of Fusarium graminearum. Therefore, the specific occurrence of A-to-I mRNA editing during the sexual reproductive stage not only avoids the trade-off between survival and reproduction caused by editing itself, but also resolves the trade-off caused by antagonistic pleiotropy of genes, thus promoting the optimization of fungal survival and reproduction.
The origin and evolution of the A-to-I mRNA editing mechanism in fungi. The homologs of the mRNA editing activator Ame1 are widely present in the fungal kingdom and have undergone multiple duplication and horizontal gene transfer events in different fungal lineages. Specifically, a gene duplication event occurred in the most recent common ancestor of the Sordariomycetes and Leotiomycetes fungi. One copy of this duplication, during the evolution of the Sordariomycetes fungi, gained the ability to interact with Tad3 through rapid mutations, ultimately evolving into the mRNA editing activator Ame1.
The fungal A-to-I mRNA editing system has broad application prospects. The study shows that the fungal A-to-I mRNA editing system has widespread applicability and can be used in the design and development of gene editing tools, especially for gene therapy to repair human genetic diseases. Additionally, the A-to-I mRNA editing enzyme complex will be an excellent target for controlling fungal diseases.
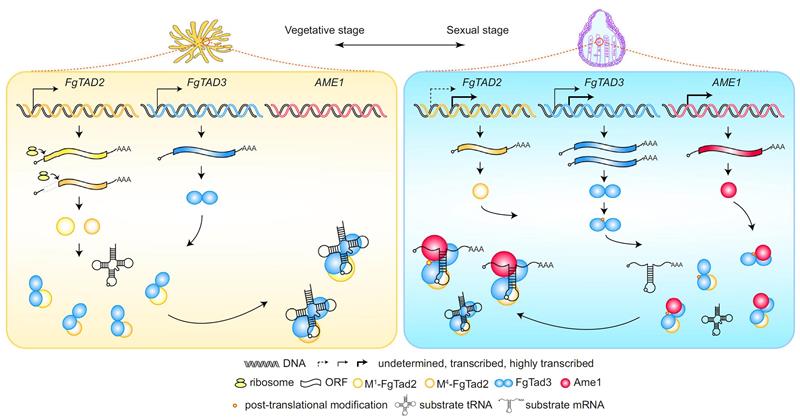
The working principle and regulatory mechanism of the A-to-I mRNA editing enzyme complex in fungi
Prof. Liu Huiquan from our college is the corresponding author of this paper, and Ph.D students Feng Chanjing (now a postdoctoral researcher), Xin Kaiyun (now a postdoctoral researcher), Du Yanfei, and Zou Jingwen are the co-first authors. Prof. Kang Zhensheng, Prof. Wang Xiaojie, Researcher Jiang Cong, and Associate Prof. Wang Qinhu from our college, as well as Associate Prof. Huang Weiwei from the College of Life Sciences, have also made significant contributions to this research. The study was funded by the National Key Research and Development Program, the National Natural Science Foundation of China, and the Postdoctoral Innovative Talents Support Program.
Original link:
The research group was invited to share this scientific discovery on Springer Nature Research Communities:
Unveiling the Mystery of Fungal RNA Editing Machinery: A Story of Unexpected Discoveries