On March 14th, the research team on functional genomics of crop pathogenic fungi in our College published an online research paper entitled 'Experimental evidence for the functional importance and adaptive advantage of A-to-I RNA editing in fungi' in the Proceedings of the National Academy of Sciences (PNAS) of the United States. Doctoral student Kaiyun Xin is the first author of the paper, and Professor Huiquan Liu is the corresponding author.
Among the known types of RNA modifications, only a few can alter the coding ability of RNA sequences, and these modifications are called RNA editing. Most types of RNA editing only exist in organelles (such as C-to-U RNA editing in plant mitochondria and chloroplasts), and only a few occur in RNA encoded by the nuclear genome, of which the most famous and extensively studied is A-to-I RNA editing. A-to-I RNA editing was first reported in animals in 1988 and was not reported in species outside of animals until 2016 by the research team on functional genomics of crop pathogenic fungi. A-to-I RNA editing converts adenosine (A) in RNA molecules to inosine (I) through enzymatic deamination, and inosine is recognized as guanosine (G) by various cellular processes. Therefore, A-to-I RNA editing is equivalent to an A-to-G mutation on RNA, and if editing occurs in the first two positions of a codon, it can lead to changes in the protein sequence.
Due to the fact that RNA editing has a certain editing level at specific sites, a single cell can simultaneously express a certain proportion of unedited and edited RNA/proteins. Therefore, it is generally assumed that RNA editing has the advantage of increasing cellular protein diversity and improving species adaptability. However, this hypothesis has been controversial due to the lack of supporting experimental evidence. The research team systematically studied the functional significance of A-to-I RNA editing sites and their associated genes in Fusarium graminearum through gene knockout and site-directed mutagenesis, and found that the editing sites on the CME5 and CME11 genes play important regulatory roles in the sexual reproduction of F. graminearum (Figure 1). The strains that only expressed edited CME5 protein had normal sexual reproduction, while the strains that only expressed unedited CME5 protein had severe defects in ascus and ascospore formation, similar to the knockout mutants (Figure 1A), indicating that RNA editing of CME5 plays an important role in regulating sexual development. At the CME5 editing site, the unedited amino acid residue (R) is conserved in the ascomycete fungi, while the edited amino acid residue (G) is rare (Figure 1B), indicating that producing both R and G proteins through RNA editing at this site may have higher evolutionary adaptability than expressing only one edited G protein through genomic mutation. More importantly, the study found that strains that only expressed the unedited (T residue) or edited (A residue) form of CME11 protein had significant defects in ascospore formation, while strains that simultaneously expressed both forms of CME11 protein showed normal performance (Figure 1C), indicating that the editing site on CME11 is not only important but also has "heterozygote advantage", that is, expressing both forms of protein simultaneously has greater advantage than expressing any one form of protein alone. Further studies revealed that the editing site on CME11 protein is likely to be phosphorylated, and the T>A amino acid change caused by A-to-I RNA editing has a mimicking effect of dephosphorylation (Figure 1D), indicating that RNA editing may regulate sexual development by modulating the phosphorylation level at this site, revealing the potential mechanism of the formation of RNA editing "heterozygote advantage”.
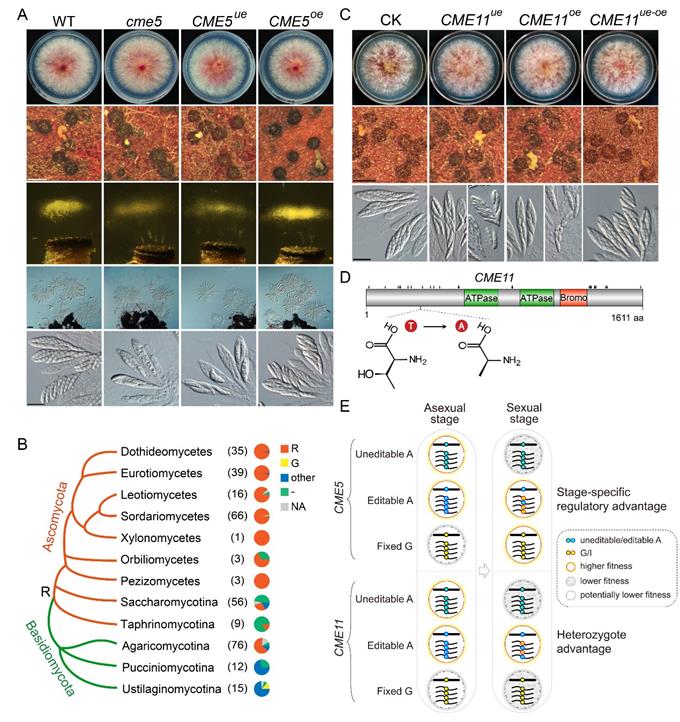
Figure 1. Functional significance and adaptive advantages of RNA editing sites on CME5 and CME11.
Professor Cong Jiang, Associate Professor Qinhu Wang, and other graduate students of the team made important contributions to this work. This research was supported by the National Key R&D Program and the National Natural Science Foundation of China. We would like to thank the State Key Laboratory of Crop Stress Biology in Arid Areas for their support of the research platform.
Original article: https://www.pnas.org/doi/10.1073/pnas.2219029120